19 Improving performance
19.1 Checking for existing solutions
Q1: What are faster alternatives to lm? Which are specifically designed to work with larger datasets?
A: The CRAN task view for high-performance computing provides many recommendations. For this question, we are most interested in the section on “Large memory and out-of-memory data.” We could for example give biglm::biglm(),27 speedglm::speedlm()28 or RcppEigen::fastLm()29 a try.
For small datasets, we observe only minor performance gains (or even a small cost):
penguins <- palmerpenguins::penguins
bench::mark(
"lm" = lm(
body_mass_g ~ bill_length_mm + species, data = penguins
) %>% coef(),
"biglm" = biglm::biglm(
body_mass_g ~ bill_length_mm + species, data = penguins
) %>% coef(),
"speedglm" = speedglm::speedlm(
body_mass_g ~ bill_length_mm + species, data = penguins
) %>% coef(),
"fastLm" = RcppEigen::fastLm(
body_mass_g ~ bill_length_mm + species, data = penguins
) %>% coef()
)
#> # A tibble: 4 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 lm 1.24ms 1.34ms 749. 908.48KB 4.13
#> 2 biglm 940.12µs 992.4µs 989. 5.43MB 6.40
#> 3 speedglm 1.6ms 1.68ms 589. 62.43MB 2.02
#> 4 fastLm 972.62µs 1.01ms 967. 3.42MB 2.03For larger datasets the selection of the appropriate method is of greater relevance:
eps <- rnorm(100000)
x1 <- rnorm(100000, 5, 3)
x2 <- rep(c("a", "b"), 50000)
y <- 7 * x1 + (x2 == "a") + eps
td <- data.frame(y = y, x1 = x1, x2 = x2, eps = eps)
bench::mark(
"lm" = lm(y ~ x1 + x2, data = td) %>% coef(),
"biglm" = biglm::biglm(y ~ x1 + x2, data = td) %>% coef(),
"speedglm" = speedglm::speedlm(y ~ x1 + x2, data = td) %>% coef(),
"fastLm" = RcppEigen::fastLm(y ~ x1 + x2, data = td) %>% coef()
)
#> # A tibble: 4 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 lm 46.3ms 48.6ms 13.4 27MB 17.3
#> 2 biglm 40.4ms 43.5ms 17.2 22.2MB 21.0
#> 3 speedglm 38.1ms 41.1ms 23.9 22.9MB 19.9
#> 4 fastLm 66.3ms 75ms 13.7 30.2MB 15.6For further speed improvements, you could install a linear algebra library optimised for your system (see ?speedglm::speedlm).
The functions of class ‘speedlm’ may speed up the fitting of LMs to large datasets. High performances can be obtained especially if R is linked against an optimized BLAS, such as ATLAS.
Tip: In case your dataset is stored in a database, you might want to check out the {modeldb} package30 which executes the linear model code in the corresponding database backend.
Q2: What package implements a version of match() that’s faster for repeated lookups? How much faster is it?
A: A web search points us to the fastmatch package.31 We compare it to base::match() and observe an impressive performance gain.
table <- 1:100000
x <- sample(table, 10000, replace = TRUE)
bench::mark(
match = match(x, table),
fastmatch = fastmatch::fmatch(x, table)
)
#> # A tibble: 2 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 match 18.9ms 19.5ms 51.3 1.46MB 2.05
#> 2 fastmatch 552.9µs 594.6µs 1671. 442.91KB 2.04Q3: List four functions (not just those in base R) that convert a string into a date time object. What are their strengths and weaknesses?
A: The usual base R way is to use the as.POSIXct() generic and create a date time object of class POSIXct and type integer.
date_ct <- as.POSIXct("2020-01-01 12:30:25")
date_ct
#> [1] "2020-01-01 12:30:25 UTC"Under the hood as.POSIXct() employs as.POSIXlt() for the character conversion. This creates a date time object of class POSIXlt and type list.
date_lt <- as.POSIXlt("2020-01-01 12:30:25")
date_lt
#> [1] "2020-01-01 12:30:25 UTC"The POSIXlt class has the advantage that it carries the individual time components as attributes. This allows to extract the time components via typical list operators.
attributes(date_lt)
#> $names
#> [1] "sec" "min" "hour" "mday" "mon" "year" "wday" "yday" "isdst"
#>
#> $class
#> [1] "POSIXlt" "POSIXt"
#>
#> $tzone
#> [1] "UTC"
date_lt$sec
#> [1] 25However, while lists may be practical, basic calculations are often faster and require less memory for objects with underlying integer type.
date_lt2 <- rep(date_lt, 10000)
date_ct2 <- rep(date_ct, 10000)
bench::mark(
date_lt2 - date_lt2,
date_ct2 - date_ct2,
date_ct2 - date_lt2
)
#> # A tibble: 3 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 date_lt2 - date_lt2 4.27ms 4.41ms 226. 1.14MB 11.5
#> 2 date_ct2 - date_ct2 89.48µs 177.61µs 5613. 195.45KB 24.9
#> 3 date_ct2 - date_lt2 2.1ms 2.29ms 437. 664.67KB 7.37as.POSIXlt() in turn uses strptime() under the hood, which creates a similar date time object.
date_str <- strptime("2020-01-01 12:30:25",
format = "%Y-%m-%d %H:%M:%S")
identical(date_lt, date_str)
#> [1] TRUEas.POSIXct() and as.POSIXlt() accept different character inputs by default (e.g. "2001-01-01 12:30" or "2001/1/1 12:30"). strptime() requires the format argument to be set explicitly, and provides a performance improvement in return.
bench::mark(
as.POSIXct = as.POSIXct("2020-01-01 12:30:25"),
as.POSIXct_format = as.POSIXct("2020-01-01 12:30:25",
format = "%Y-%m-%d %H:%M:%S"
),
strptime_fomat = strptime("2020-01-01 12:30:25",
format = "%Y-%m-%d %H:%M:%S"
)
)[1:3]
#> # A tibble: 3 x 3
#> expression min median
#> <bch:expr> <bch:tm> <bch:tm>
#> 1 as.POSIXct 95.69µs 100.4µs
#> 2 as.POSIXct_format 30.37µs 33.4µs
#> 3 strptime_fomat 9.29µs 10.3µsA fourth way is to use the converter functions from the lubridate package,32 which contains wrapper functions (for the POSIXct approach) with an intuitive syntax. (There is a slight decrease in performance though.)
library(lubridate)
ymd_hms("2013-07-24 23:55:26")
#> [1] "2013-07-24 23:55:26 UTC"
bench::mark(
as.POSIXct = as.POSIXct("2013-07-24 23:55:26", tz = "UTC"),
ymd_hms = ymd_hms("2013-07-24 23:55:26")
)[1:3]
#> # A tibble: 2 x 3
#> expression min median
#> <bch:expr> <bch:tm> <bch:tm>
#> 1 as.POSIXct 92.31µs 96.99µs
#> 2 ymd_hms 4.46ms 4.63msFor additional ways to convert characters into date time objects, have a look at the {chron}, the {anytime} and the {fasttime} packages. The {chron} package33 introduces new classes and stores times as fractions of days in the underlying double type, while it doesn’t deal with time zones and daylight savings. The {anytime} package34 aims to convert “Anything to POSIXct or Date.” The {fasttime} package35 contains only one function, fastPOSIXct().
Q4: Which packages provide the ability to compute a rolling mean?
A: A rolling mean is a useful statistic to smooth time-series, spatial and other types of data. The size of the rolling window usually determines the amount of smoothing and the number of missing values at the boundaries of the data.
The general functionality can be found in multiple packages, which vary in speed and flexibility of the computations. Here is a benchmark for several functions that all serve our purpose.
x <- 1:10
slider::slide_dbl(x, mean, .before = 1, .complete = TRUE)
#> [1] NA 1.5 2.5 3.5 4.5 5.5 6.5 7.5 8.5 9.5
bench::mark(
caTools = caTools::runmean(x, k = 2, endrule = "NA"),
data.table = data.table::frollmean(x, 2),
RcppRoll = RcppRoll::roll_mean(x, n = 2, fill = NA,
align = "right"),
slider = slider::slide_dbl(x, mean, .before = 1, .complete = TRUE),
TTR = TTR::SMA(x, 2),
zoo_apply = zoo::rollapply(x, 2, mean, fill = NA, align = "right"),
zoo_rollmean = zoo::rollmean(x, 2, fill = NA, align = "right")
)
#> # A tibble: 7 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 caTools 89.6µs 95.2µs 10414. 167.64KB 4.10
#> 2 data.table 49.9µs 55.4µs 17435. 1.42MB 6.23
#> 3 RcppRoll 47.5µs 51.1µs 19372. 39.23KB 4.08
#> 4 slider 76.2µs 84.7µs 11701. 0B 4.07
#> 5 TTR 470µs 489.6µs 2034. 1.18MB 2.02
#> 6 zoo_apply 389.4µs 421.5µs 2344. 430.22KB 4.07
#> 7 zoo_rollmean 334.6µs 370.8µs 2672. 6.42KB 4.08You may also take a look at an extensive example in the first edition of Advanced R, which demonstrates how a rolling mean function can be created.
Q5: What are the alternatives to optim()?
A: According to its description (see ?optim) optim() implements:
General-purpose optimization based on Nelder–Mead, quasi-Newton and conjugate-gradient algorithms. It includes an option for box-constrained optimization and simulated annealing.
optim() allows to optimise a function (fn) on an interval with a specific method (method = c("Nelder-Mead", "BFGS", "CG", "L-BFGS-B", "SANN", "Brent")). Many detailed examples are given in the documentation. In the simplest case, we give optim() the starting value par = 0 to calculate the minimum of a quadratic polynomial:
optim(0, function(x) x^2 - 100 * x + 50,
method = "Brent",
lower = -1e20, upper = 1e20
)
#> $par
#> [1] 50
#>
#> $value
#> [1] -2450
#>
#> $counts
#> function gradient
#> NA NA
#>
#> $convergence
#> [1] 0
#>
#> $message
#> NULLSince this solves a one-dimensional optimisation task, we could have also used stats::optimize().
optimize(function(x) x^2 - 100 * x + 50, c(-1e20, 1e20))
#> $minimum
#> [1] 50
#>
#> $objective
#> [1] -2450For more general alternatives, the appropriate choice highly depends on the type of optimisation you intend to do. The CRAN task view on optimisation and mathematical modelling can serve as a useful reference. Here are a couple of examples:
-
{optimx}36 extends theoptim()function with the same syntax but moremethodchoices. -
{RcppNumerical}37 wraps several open source libraries for numerical computing (written in C++) and integrates them with R via Rcpp. -
{DEoptim}38 provides a global optimiser based on the Differential Evolution algorithm.
19.2 Doing as little as possible
Q1: What’s the difference between rowSums() and .rowSums()?
A: When we inspect the source code of the user-facing rowSums(), we see that it is designed as a wrapper around .rowSums() with some input validation, conversions and handling of complex numbers.
rowSums
#> function (x, na.rm = FALSE, dims = 1L)
#> {
#> if (is.data.frame(x))
#> x <- as.matrix(x)
#> if (!is.array(x) || length(dn <- dim(x)) < 2L)
#> stop("'x' must be an array of at least two dimensions")
#> if (dims < 1L || dims > length(dn) - 1L)
#> stop("invalid 'dims'")
#> p <- prod(dn[-(id <- seq_len(dims))])
#> dn <- dn[id]
#> z <- if (is.complex(x))
#> .Internal(rowSums(Re(x), prod(dn), p, na.rm)) + (0+1i) *
#> .Internal(rowSums(Im(x), prod(dn), p, na.rm))
#> else .Internal(rowSums(x, prod(dn), p, na.rm))
#> if (length(dn) > 1L) {
#> dim(z) <- dn
#> dimnames(z) <- dimnames(x)[id]
#> }
#> else names(z) <- dimnames(x)[[1L]]
#> z
#> }
#> <bytecode: 0x7fdc052ed7b0>
#> <environment: namespace:base>.rowSums() calls an internal function, which is built into the R interpreter. These compiled functions can be very fast.
.rowSums
#> function (x, m, n, na.rm = FALSE)
#> .Internal(rowSums(x, m, n, na.rm))
#> <bytecode: 0x7fdc05a1fe40>
#> <environment: namespace:base>However, as our benchmark reveals almost identical computing times, we prefer the safer variant over the internal function for this case.
m <- matrix(rnorm(1e6), nrow = 1000)
bench::mark(
rowSums(m),
.rowSums(m, 1000, 1000)
)
#> # A tibble: 2 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 rowSums(m) 1.67ms 1.79ms 559. 7.86KB 0
#> 2 .rowSums(m, 1000, 1000) 1.62ms 1.77ms 565. 7.86KB 0Q2: Make a faster version of chisq.test() that only computes the chi-square test statistic when the input is two numeric vectors with no missing values. You can try simplifying chisq.test() or by coding from the mathematical definition.
A: We aim to speed up our reimplementation of chisq.test() by doing less.
chisq.test2 <- function(x, y) {
m <- rbind(x, y)
margin1 <- rowSums(m)
margin2 <- colSums(m)
n <- sum(m)
me <- tcrossprod(margin1, margin2) / n
x_stat <- sum((m - me)^2 / me)
df <- (length(margin1) - 1) * (length(margin2) - 1)
p.value <- pchisq(x_stat, df = df, lower.tail = FALSE)
list(x_stat = x_stat, df = df, p.value = p.value)
}We check if our new implementation returns the same results and benchmark it afterwards.
a <- 21:25
b <- seq(21, 29, 2)
m <- cbind(a, b)
chisq.test(m) %>% print(digits=5)
#>
#> Pearson's Chi-squared test
#>
#> data: m
#> X-squared = 0.162, df = 4, p-value = 1
chisq.test2(a, b)
#> $x_stat
#> [1] 0.162
#>
#> $df
#> [1] 4
#>
#> $p.value
#> [1] 0.997
bench::mark(
chisq.test(m),
chisq.test2(a, b),
check = FALSE
)
#> # A tibble: 2 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 chisq.test(m) 106.2µs 113µs 8686. 0B 2.03
#> 2 chisq.test2(a, b) 31.8µs 34.1µs 28932. 0B 5.79Q3: Can you make a faster version of table() for the case of an input of two integer vectors with no missing values? Can you use it to speed up your chi-square test?
A: When analysing the source code of table() we aim to omit everything unnecessary and extract the main building blocks. We observe that table() is powered by tabulate() which is a very fast counting function. This leaves us with the challenge to compute the pre-processing as performant as possible.
First, we calculate the dimensions and names of the output table. Then we use fastmatch::fmatch() to map the elements of each vector to their position within the vector itself (i.e. the smallest value is mapped to 1L, the second smallest value to 2L, etc.). Following the logic within table() we combine and shift these values to create a mapping of the integer pairs in our data to the index of the output table. After applying these lookups tabulate() counts the values and returns an integer vector with counts for each position in the table. As a last step, we reuse the code from table() to assign the correct dimension and class.
table2 <- function(a, b){
a_s <- sort(unique(a))
b_s <- sort(unique(b))
a_l <- length(a_s)
b_l <- length(b_s)
dims <- c(a_l, b_l)
pr <- a_l * b_l
dn <- list(a = a_s, b = b_s)
bin <- fastmatch::fmatch(a, a_s) +
a_l * fastmatch::fmatch(b, b_s) - a_l
y <- tabulate(bin, pr)
y <- array(y, dim = dims, dimnames = dn)
class(y) <- "table"
y
}
a <- sample(100, 10000, TRUE)
b <- sample(100, 10000, TRUE)
bench::mark(
table(a, b),
table2(a, b)
)
#> # A tibble: 2 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 table(a, b) 1.74ms 1.9ms 528. 1.19MB 17.0
#> 2 table2(a, b) 637.01µs 821µs 1198. 694.48KB 16.2Since we didn’t use table() in our chisq.test2()-implementation, we cannot benefit from the slight performance gain from table2().
19.3 Vectorise
Q1: The density functions, e.g. dnorm(), have a common interface. Which arguments are vectorised over? What does rnorm(10, mean = 10:1) do?
A: We can get an overview of the interface of these functions via ?dnorm:
dnorm(x, mean = 0, sd = 1, log = FALSE)
pnorm(q, mean = 0, sd = 1, lower.tail = TRUE, log.p = FALSE)
qnorm(p, mean = 0, sd = 1, lower.tail = TRUE, log.p = FALSE)
rnorm(n, mean = 0, sd = 1)These functions are vectorised over their numeric arguments, which includes the first argument (x, q, p, n) as well as mean and sd.
rnorm(10, mean = 10:1) generates ten random numbers from different normal distributions. These normal distributions differ in their means. The first has mean 10, the second mean 9, the third mean 8 and so on.
Q2: Compare the speed of apply(x, 1, sum) with rowSums(x) for varying sizes of x.
A: We compare the two functions for square matrices of increasing size:
rowsums <- bench::press(
p = seq(500, 5000, length.out = 10),
{
mat <- tcrossprod(rnorm(p), rnorm(p))
bench::mark(
rowSums = rowSums(mat),
apply = apply(mat, 1, sum)
)
}
)
#> Running with:
#> p
#> 1 500
#> 2 1000
#> 3 1500
#> 4 2000
#> 5 2500
#> 6 3000
#> 7 3500
#> 8 4000
#> 9 4500
#> 10 5000
library(ggplot2)
rowsums %>%
summary() %>%
dplyr::mutate(Approach = as.character(expression)) %>%
ggplot(
aes(p, median, color = Approach, group = Approach)) +
geom_point() +
geom_line() +
labs(x = "Number of Rows and Columns",
y = "Median (s)") +
theme(legend.position = "top")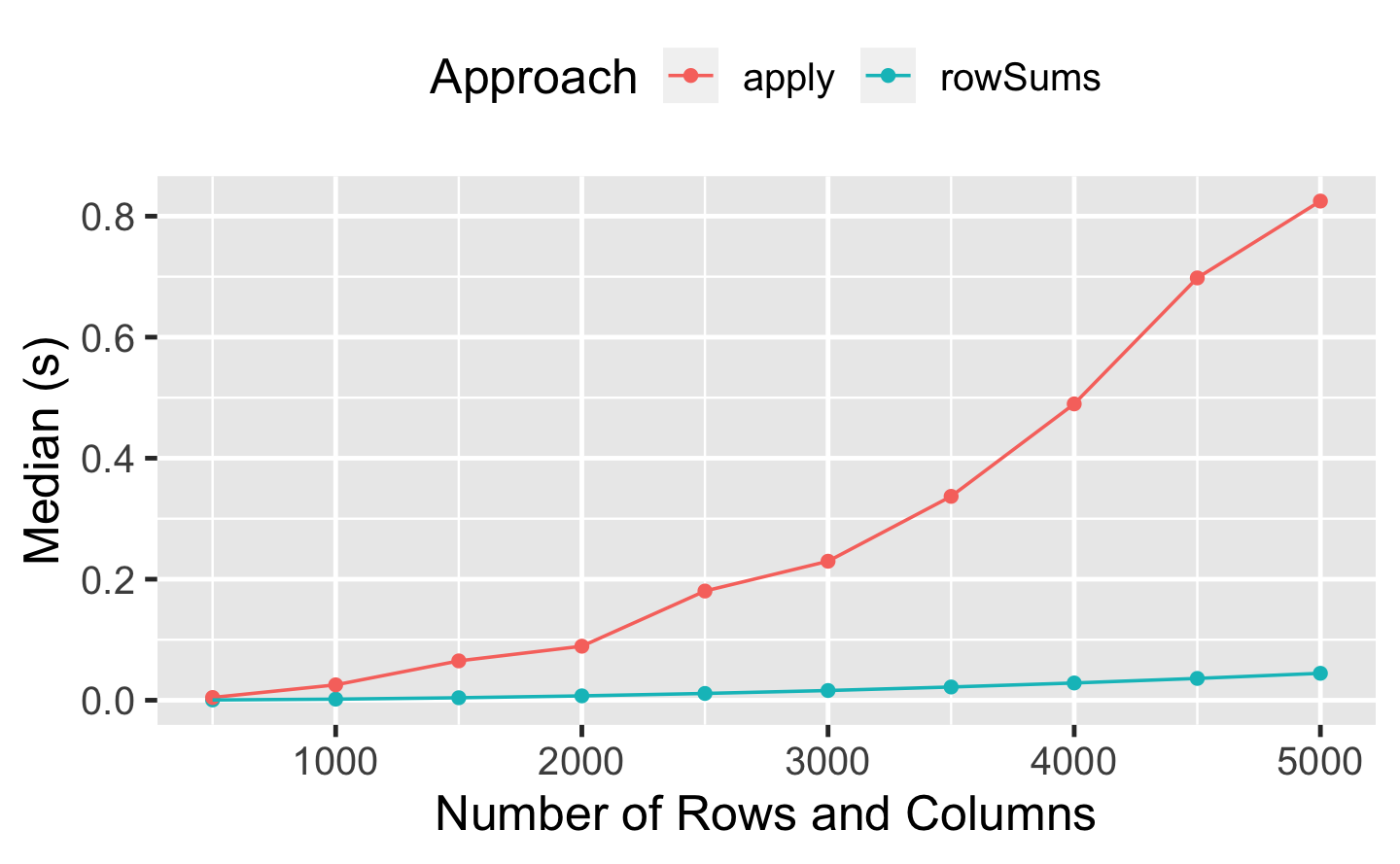
We can see that the difference in performance is negligible for small matrices but becomes more and more relevant as the size of the data increases. apply() is a very versatile tool, but it’s not “vectorised for performance” and not as optimised as rowSums().
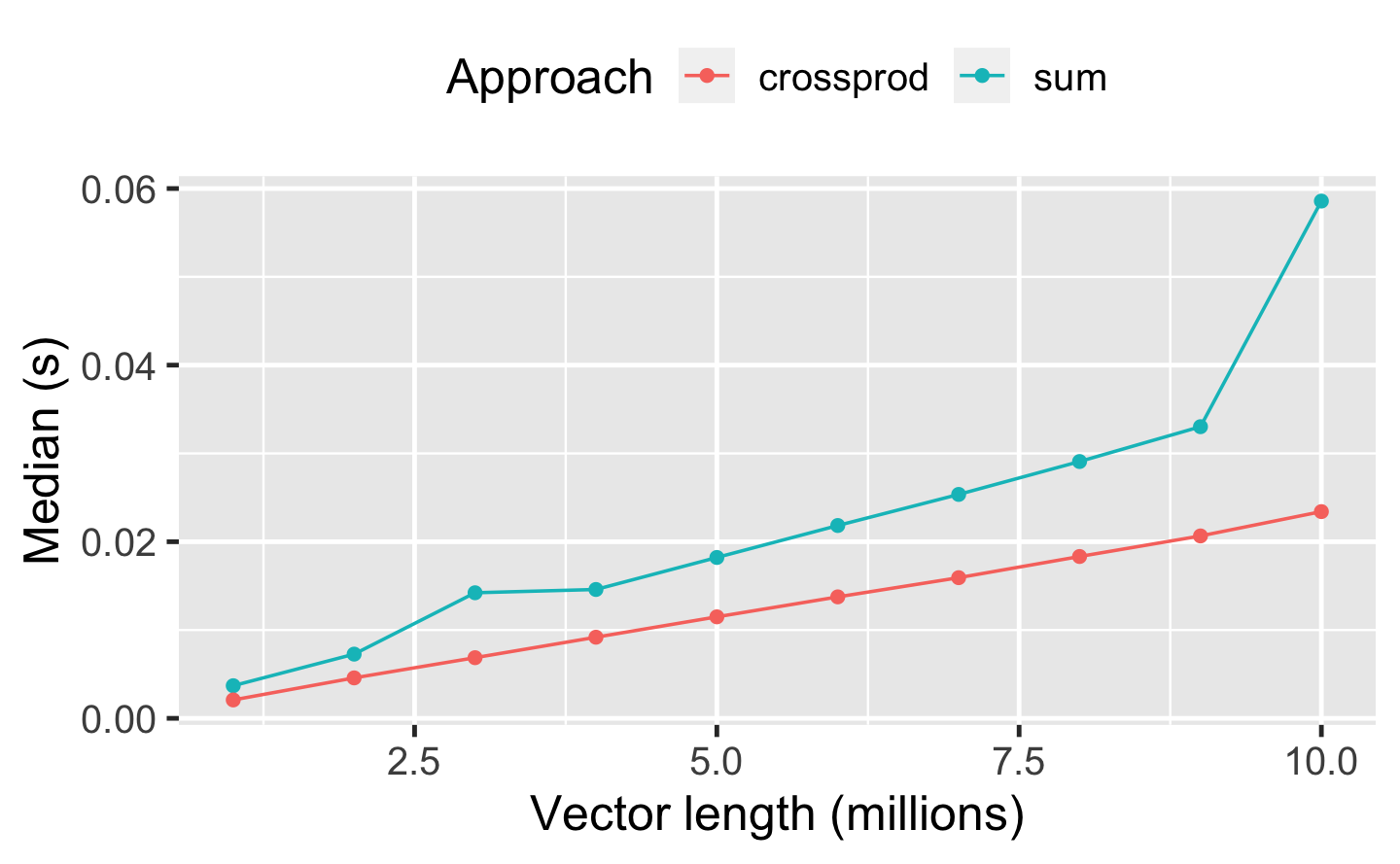
Q3: How can you use crossprod() to compute a weighted sum? How much faster is it than the naive sum(x * w)?
A: We can hand the vectors to crossprod(), which converts them to row- and column-vectors and then multiplies these. The result is the dot product, which corresponds to a weighted sum.
A benchmark of both approaches for different vector lengths indicates that the crossprod() variant is almost twice as fast as sum(x * w).
weightedsum <- bench::press(
n = 1:10,
{
x <- rnorm(n * 1e6)
bench::mark(
sum = sum(x * x),
crossprod = crossprod(x, x)[[1]]
)
}
)
#> Running with:
#> n
#> 1 1
#> 2 2
#> 3 3
#> 4 4
#> 5 5
#> 6 6
#> 7 7
#> 8 8
#> 9 9
#> 10 10
weightedsum %>%
summary() %>%
dplyr::mutate(Approach = as.character(expression)) %>%
ggplot(aes(n, median, color = Approach, group = Approach)) +
geom_point() +
geom_line() +
labs(x = "Vector length (millions)",
y = "Median (s)") +
theme(legend.position = "top")